|
产品名称 |
MEG-01人成巨核细胞白血病细胞 |
|
货号 |
ZQ0431 |
|
产品介绍 |
MEG-01细胞源自一位CML患者成巨核细胞转换期的骨髓细胞。细胞质因子Ⅷ和表面球蛋白Ⅱb/Ⅲa、高碘酸-Schiff(PAS)反应;α醋酸萘酯酶和酸性磷酸酶阳性;髓过氧化物酶、α-丁酸萘酯酶、氯化醋酸AS-D萘苯酚酯酶和碱性磷酸酶阴性。用单克隆抗体BA-1(抗B细胞、粒性白细胞),HPL-3(抗球蛋白Ⅱb/Ⅲa)和20.3(抗单核细胞、血小板)染色成阳性,其他淋巴和骨髓类抗体成阴性。 |
|
种属 |
人 |
|
性别/年龄 |
男性/55岁 |
|
组织 |
骨; 骨髓 |
|
疾病 |
急变期慢性粒细胞白血病 |
|
生物安全等级 |
BSL-1 |
|
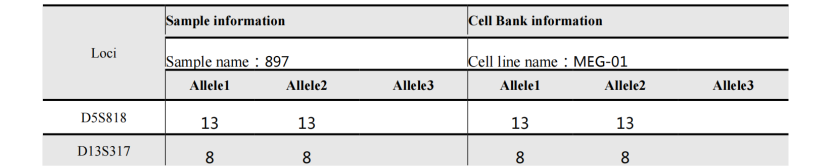
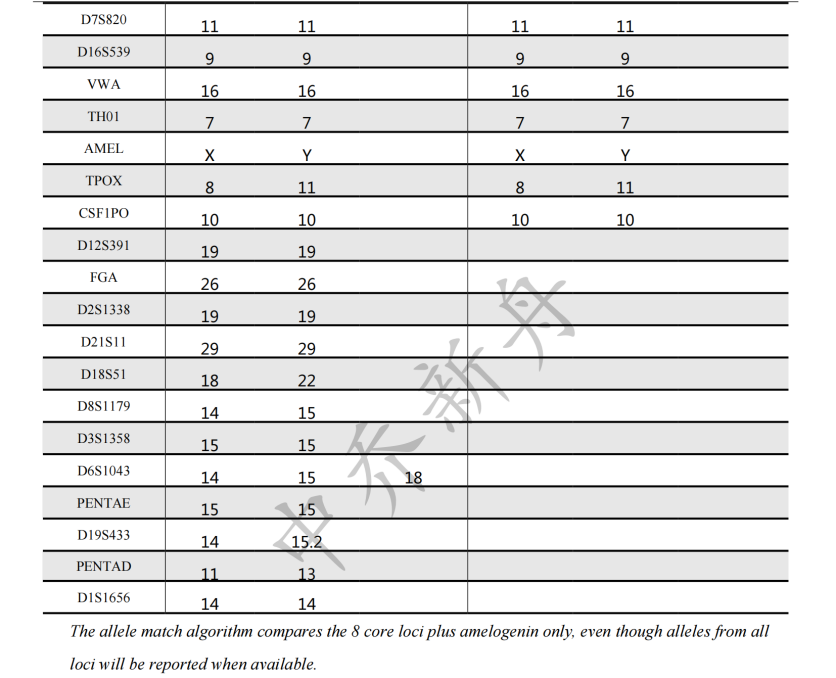
STR位点信息 |
Amelogenin X,Y CSF1PO 10 D2S1338 19 D3S1358 15 D5S818 13 D7S820 11 D8S1179 14,15 D13S317 8 D16S539 9 D18S51 18,22 D19S433 14,16 D21S11 29 FGA 26 Penta D 11,13 Penta E 15 TH01 7 TPOX 8,11 vWA 16 |
|
细胞类型 |
巨核细胞 |
|
形态学 |
淋巴母细胞 |
|
生长方式 |
混合:粘附和悬浮 |
|
倍增时间 |
36-48 hours (PubMed=2998511); ~35 hours (DSMZ=ACC-364) |
|
培养基和添加剂 |
RPMI-1640(中乔新舟 货号:ZQ-200)+20%胎牛血清(中乔新舟 货号:ZQ0500)+1%双抗(品牌:中乔新舟 货号:CSP006) |
|
推荐完全培养基货号 |
|
|
培养条件 |
95%空气,5%二氧化碳;37℃ |
|
基因表达 |
Amelogenin: X,Y CSF1PO: 10 D13S317: 8 D16S539: 9 D5S818: 13 D7S820: 11 THO1: 7 TPOX: 8,11 vWA: 16 |
|
保藏机构 |
ATCC; CRL-2021BCRC; 60238BCRJ; 0262DSMZ; ACC-364 |
|
供应限制 |
仅供科研使用 |
|
货号 |
ZQ0431 |
|
发货规格 |
活细胞:T25培养瓶*1瓶或者1ml 冻存管*2支(细胞量约为1x10^6 Cells/Vial)二选一 |
|
发货形式 |
活细胞:常温运输;冻存管:干冰运输 |
|
储存温度 |
活细胞:培养箱;冻存管:液氮罐 |
|
产地 |
中国 |
|
供应限制 |
仅供科研使用 |
PubMed=2998511; DOI=10.1182/blood.V66.6.1384.1384
Ogura M., Morishima Y., Ohno R., Kato Y., Hirabayashi N., Nagura H., Saito H.
Establishment of a novel human megakaryoblastic leukemia cell line, MEG-01, with positive Philadelphia chromosome.
Blood 66:1384-1392(1985)
PubMed=3332852; DOI=10.1016/S0950-3536(87)80037-9
Keating A.
Ph positive CML cell lines.
Baillieres Clin. Haematol. 1:1021-1029(1987)
PubMed=1742484; DOI=10.1182/blood.V78.12.3168.3168
Murate T., Hotta T., Tsushita K., Suzuki M., Yoshida T., Saga S., Saito H., Yoshida S.
Aphidicolin, an inhibitor of DNA replication, blocks the TPA-induced differentiation of a human megakaryoblastic cell line, MEG-O1.
Blood 78:3168-3177(1991)
PubMed=9738977; DOI=10.1111/j.1349-7006.1998.tb03275.x
Takizawa J., Suzuki R., Kuroda H., Utsunomiya A., Kagami Y., Joh T., Aizawa Y., Ueda R., Seto M.
Expression of the TCL1 gene at 14q32 in B-cell malignancies but not in adult T-cell leukemia.
Jpn. J. Cancer Res. 89:712-718(1998)
PubMed=10071072; DOI=10.1016/S0145-2126(98)00171-4
Drexler H.G., MacLeod R.A.F., Uphoff C.C.
Leukemia cell lines: in vitro models for the study of Philadelphia chromosome-positive leukemia.
Leuk. Res. 23:207-215(1999)
PubMed=10576511; DOI=10.1016/s0145-2126(99)00131-9
Uphoff C.C., Habig S., Fombonne S., Matsuo Y., Drexler H.G.
ABL-BCR expression in BCR-ABL-positive human leukemia cell lines.
Leuk. Res. 23:1055-1060(1999)
DOI=10.1016/B978-0-12-221970-2.50457-5
Drexler H.G.
The leukemia-lymphoma cell line factsbook.
(In) ISBN 9780122219702; pp.1-733; Academic Press; London (2001)
PubMed=15843827; DOI=10.1038/sj.leu.2403749
Andersson A., Eden P., Lindgren D., Nilsson J., Lassen C., Heldrup J., Fontes M., Borg A., Mitelman F., Johansson B., Hoglund M., Fioretos T.
Gene expression profiling of leukemic cell lines reveals conserved molecular signatures among subtypes with specific genetic aberrations.
Leukemia 19:1042-1050(2005)
PubMed=20164919; DOI=10.1038/nature08768
Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B.-Y., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F.-T., Campbell P.J., Futreal P.A., Stratton M.R.
Signatures of mutation and selection in the cancer genome.
Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458
Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A.
A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers.
Cancer Res. 70:2158-2164(2010)
PubMed=20809971; DOI=10.1186/1755-8166-3-15
Virgili A., Nacheva E.
Genomic amplification of BCR/ABL1 and a region downstream of ABL1 in chronic myeloid leukaemia: a FISH mapping study of CML patients and cell lines.
Mol. Cytogenet. 3:15.1-15.12(2010)
PubMed=22460905; DOI=10.1038/nature11003
Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.-Y.K., Yu J.-J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N.-X., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M.L., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A.
The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity.
Nature 483:603-607(2012)
PubMed=26700326; DOI=10.1016/j.exphem.2015.11.011
Inoue D., Matsumoto M., Nagase R., Saika M., Fujino T., Nakayama K.I., Kitamura T.
Truncation mutants of ASXL1 observed in myeloid malignancies are expressed at detectable protein levels.
Exp. Hematol. 44:172-176(2016)
PubMed=27277069; DOI=10.1160/TH15-11-0891
Wright J.R., Amisten S., Goodall A.H., Mahaut-Smith M.P.
Transcriptomic analysis of the ion channelome of human platelets and megakaryocytic cell lines.
Thromb. Haemost. 116:272-284(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017
Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X.-M., Egan R.K., Liu Q.-S., Mironenko T., Mitropoulos X., Richardson L., Wang J.-H., Zhang T.-H., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J.
A landscape of pharmacogenomic interactions in cancer.
Cell 166:740-754(2016)
PubMed=30285677; DOI=10.1186/s12885-018-4840-5
Tan K.-T., Ding L.-W., Sun Q.-Y., Lao Z.-T., Chien W., Ren X., Xiao J.-F., Loh X.-Y., Xu L., Lill M., Mayakonda A., Lin D.-C., Yang H., Koeffler H.P.
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines.
BMC Cancer 18:940.1-940.13(2018)
PubMed=30332548; DOI=10.1080/09537104.2018.1528344
Dhenge A., Kuhikar R., Kale V., Limaye L.S.
Regulation of differentiation of MEG01 to megakaryocytes and platelet-like particles by valproic acid through Notch3 mediated actin polymerization.
Platelets 30:780-795(2019)
PubMed=30629668; DOI=10.1371/journal.pone.0210404
Uphoff C.C., Pommerenke C., Denkmann S.A., Drexler H.G.
Screening human cell lines for viral infections applying RNA-Seq data analysis.
PLoS ONE 14:E0210404-E0210404(2019)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747
Dutil J., Chen Z.-H., Monteiro A.N.A., Teer J.K., Eschrich S.A.
An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines.
Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3
Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J.G., Gelfand E.T., Bielski C.M., Li H.-X., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y.-L., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R.
Next-generation characterization of the Cancer Cell Line Encyclopedia.
Nature 569:503-508(2019)
PubMed=32180119; DOI=10.1007/s12185-020-02853-6
Saito H., Hayakawa M., Kamoshita N., Yasumoto A., Suzuki-Inoue K., Yatomi Y., Ohmori T.
Establishment of a megakaryoblastic cell line for conventional assessment of platelet calcium signaling.
Int. J. Hematol. 111:786-794(2020)
PubMed=35839778; DOI=10.1016/j.ccell.2022.06.010
Goncalves E., Poulos R.C., Cai Z.-X., Barthorpe S., Manda S.S., Lucas N., Beck A., Bucio-Noble D., Dausmann M., Hall C., Hecker M., Koh J., Lightfoot H., Mahboob S., Mali I., Morris J., Richardson L., Seneviratne A.J., Shepherd R., Sykes E., Thomas F., Valentini S., Williams S.G., Wu Y.-X., Xavier D., MacKenzie K.L., Hains P.G., Tully B., Robinson P.J., Zhong Q., Garnett M.J., Reddel R.R.
Pan-cancer proteomic map of 949 human cell lines.
Cancer Cell 40:835-849.e8(2022)

 上海中乔新舟生物科技有限公司
上海中乔新舟生物科技有限公司